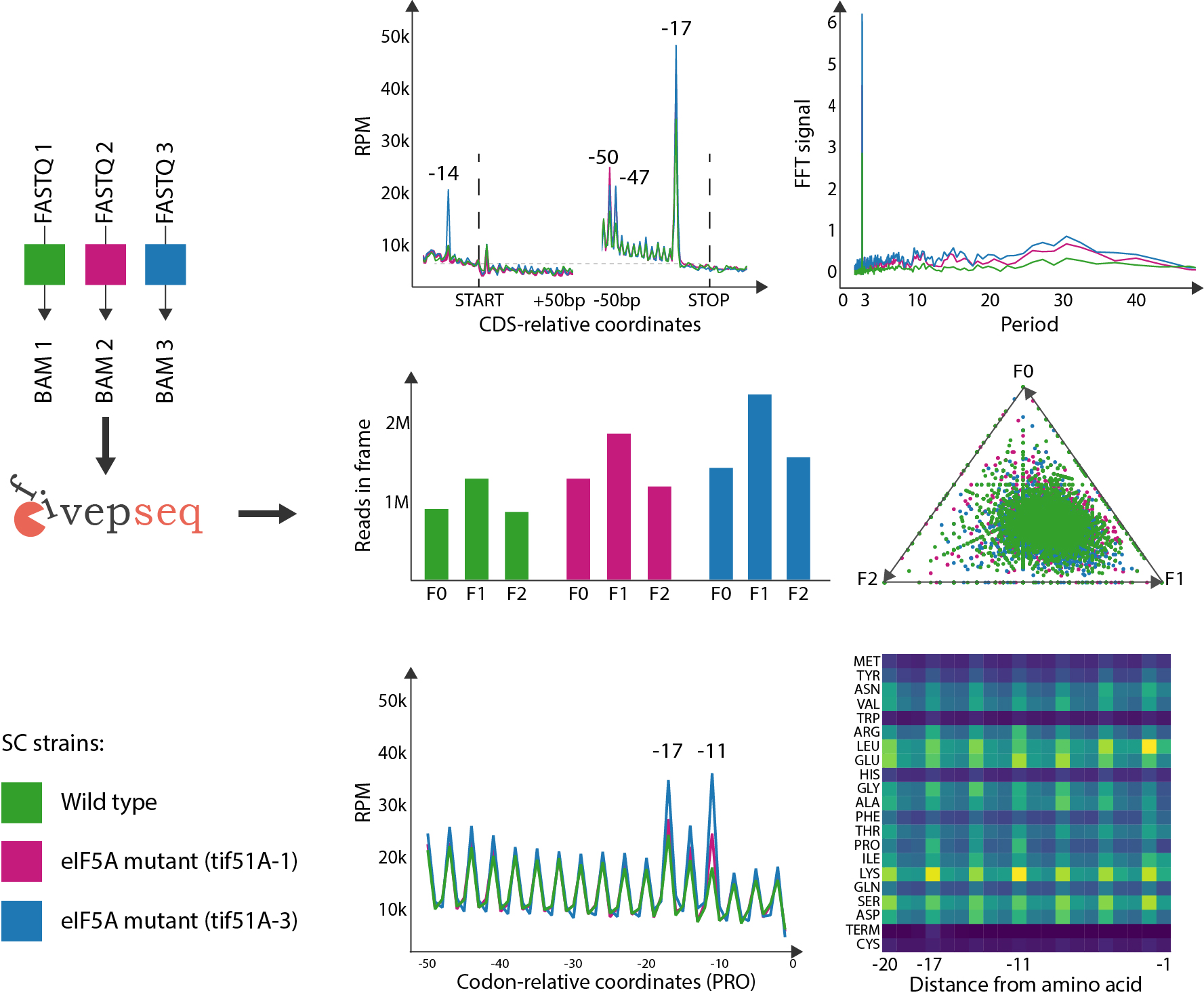
Fivepseq is a software package for analysis of 5′ endpoints distribution in RNA sequencing datasets. This is particularly useful for techniques that capture 5′ monophosphorylated RNAs, such as 5PSeq, PARE-seq or GMUC. It may also be useful for ribosome profiling datasets and alike.The main workflow of fivepseq is intended for downstream analysis of alignment files to describe the distribution of 5′ endpoints of reads relative to translation start and stop sites, as well as relative to amino acids or codons. It also computes frame preference of 5′ endpoint distribution and captures periodicity patterns.
The output of fivepseq is in the form of html report files, as well as text files and image files in portable network graphic and vector graphics formats.
Demo reports
Fivepseq was originally designed for analysis of 5′ monophosphorylated mRNA degradation intermediates generated by the 5PSeq technique. However, it can also be applied to other RNA-seq datasets, where 5′ endpoints can be used for knowledge inference, e.g. PARE, GMUCT and ribosome profiling. Below are links to example fivepseq reports from publicly available datasets for three different techniques and organisms:
Saccharomyces Cerevisiae eIF5A mutants, 5PSeq
Downloads and installation
Fivepseq source code is deposited at github, and the package along with the user guide are available at PyPI. Please, see the User manual for detailed instructions on installation and usage.
Citation
Improved computational analysis of ribosome dynamics from 5′P degradome data using fivepseq. Lilit Nersisyan, Maria Ropat & Vicent Pelechano. NAR Genomics and Bioinformatics, Volume 2, Issue 4, December 2020, lqaa099, https://doi.org/10.1093/nargab/lqaa099
Quick Install
pip install --upgrade numpy pysam cython # Install dependencies with pip
pip install plastid
git clone https://github.com/lilit-nersisyan/fivepseq # Git clone
python setup.py install # Install from fivepseq directory
fivepseq --help # See usage
conda install phantomjs selenium pillow # Install dependencies for image export